About Me
A central question in evolutionary biology is how organisms evolve highly derived and novel morphologies. I am interested in researching what changes to conserved developmental genes lead to the evolution of divergent morphologies. My background is in evolutionary genetics, specializing in developmental evolution, Hox cluster evolution, and the evolution of osteological features.
My interest in evolutionary biology began as an undergraduate student, where I worked on a research project at the California Academy of Science looking at cryptic speciation within Amolops frogs from Myanmar. This led me to study principles of systematics, taxonomy, and phylogenetics at the University of Kansas Biodiversity Institute & Natural History Museum for my Master’s degree.
At the University of Kansas, I studied the skeletal variation in Melanesian forest frogs and worked on a study examining the evolution of caudal fin skeletal development in zebrafish. After gaining a greater understanding of skeletal morphology during my Master’s degree, my research interests shifted towards wanting to study the genomic and genetic changes underlying skeletal evolution.
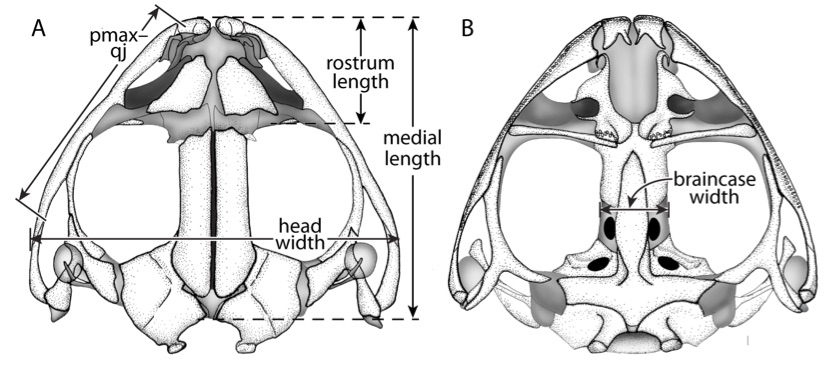
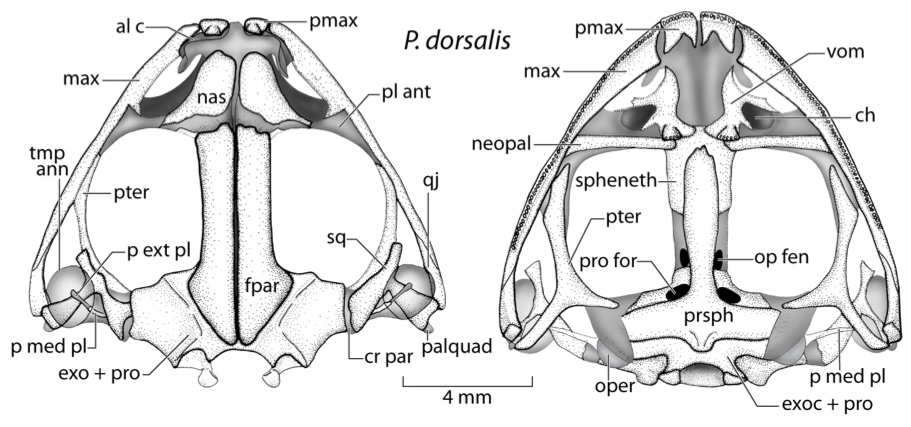
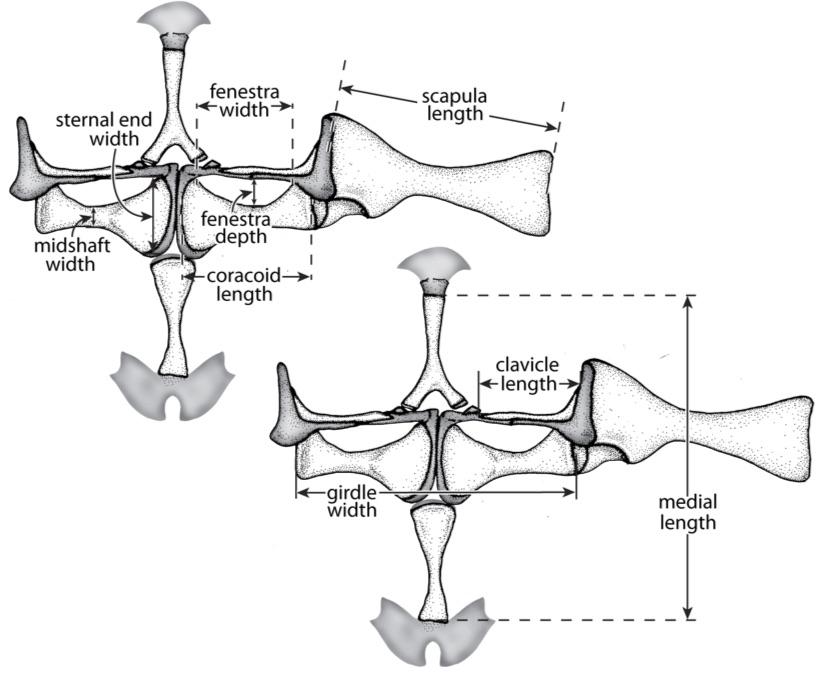
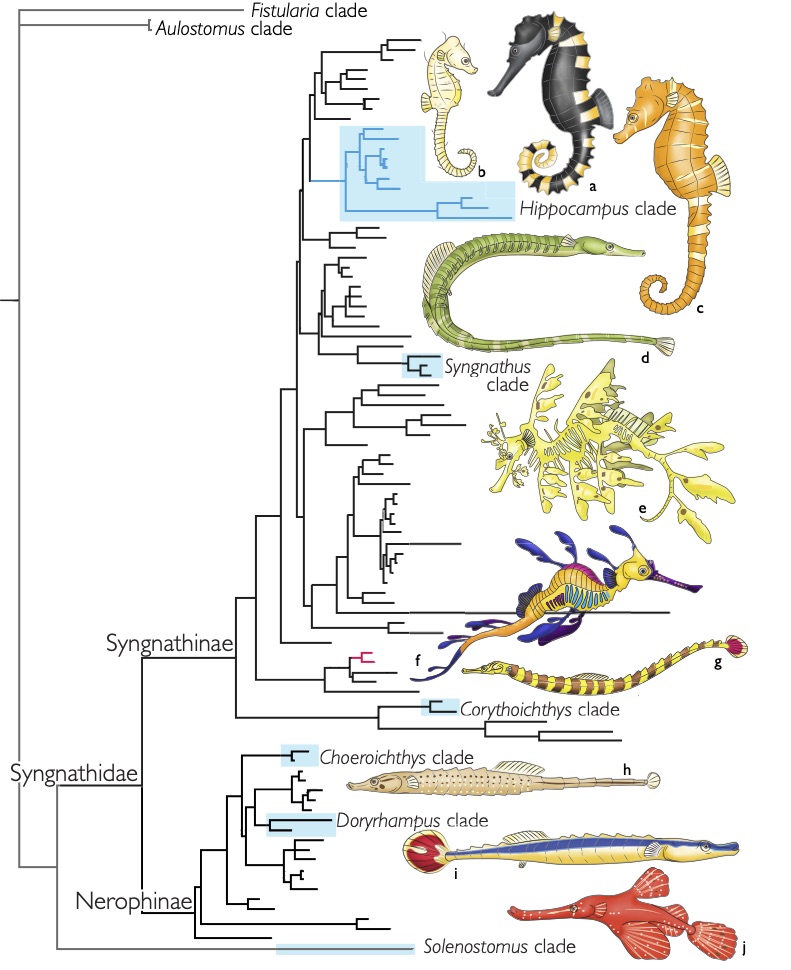
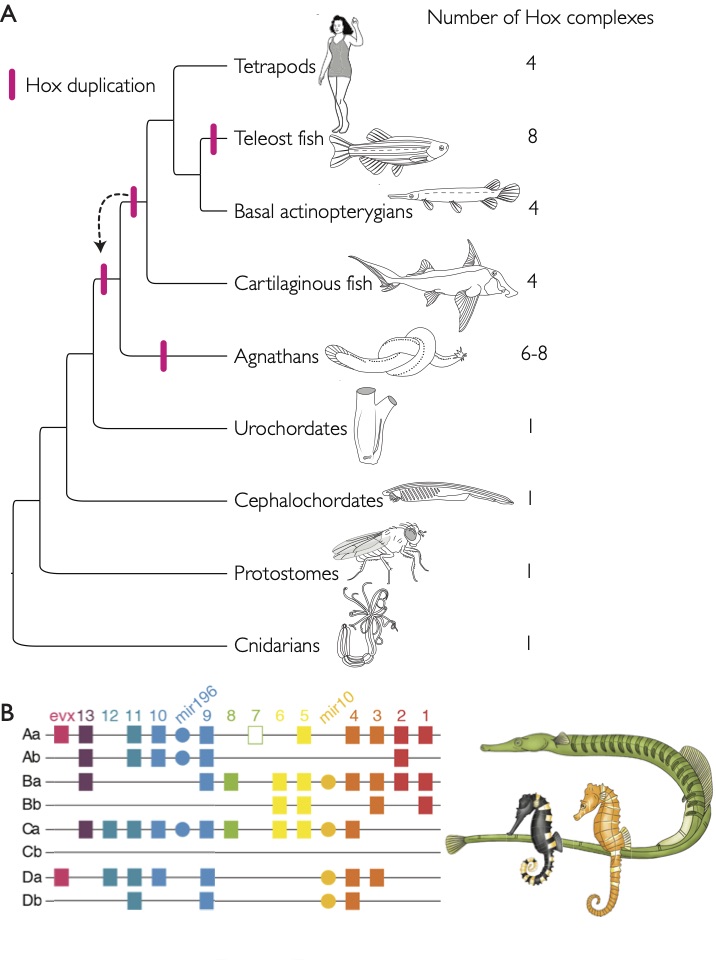
For my doctoral thesis at the University of Oregon, I researched the developmental and genomic changes in Hox genes that have contributed to the evolution of highly derived skeletal features within pipefish, seahorses, and seadragons. I contributed to the assembly and annotation of the first genome for this fascinating family of fish. I described the Hox clusters for the pipefish, which was the first annotated set of Hox genes for syngnathid fish. I developed a range of proficiencies in both molecular bench skills and bioinformatic analysis.
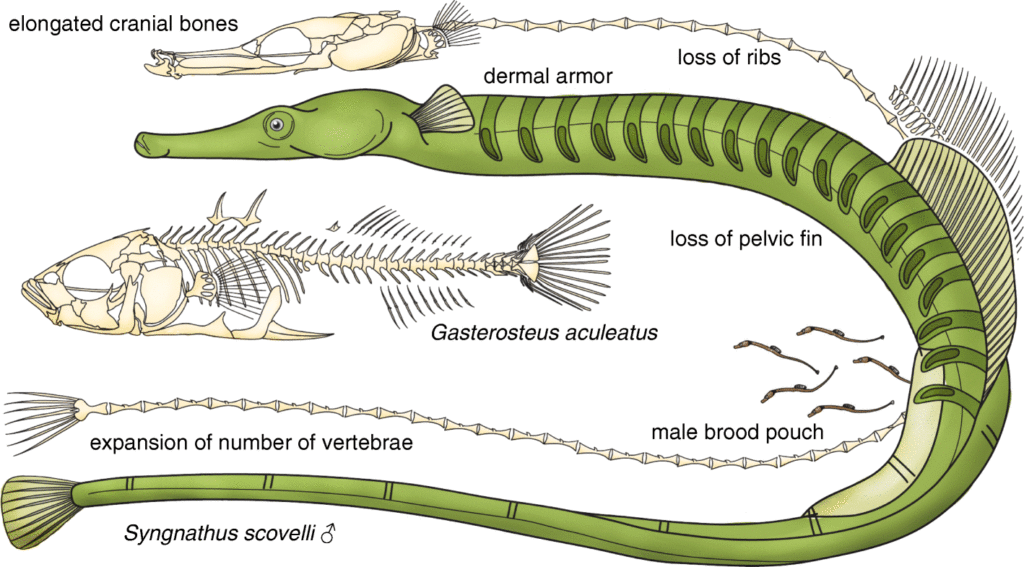
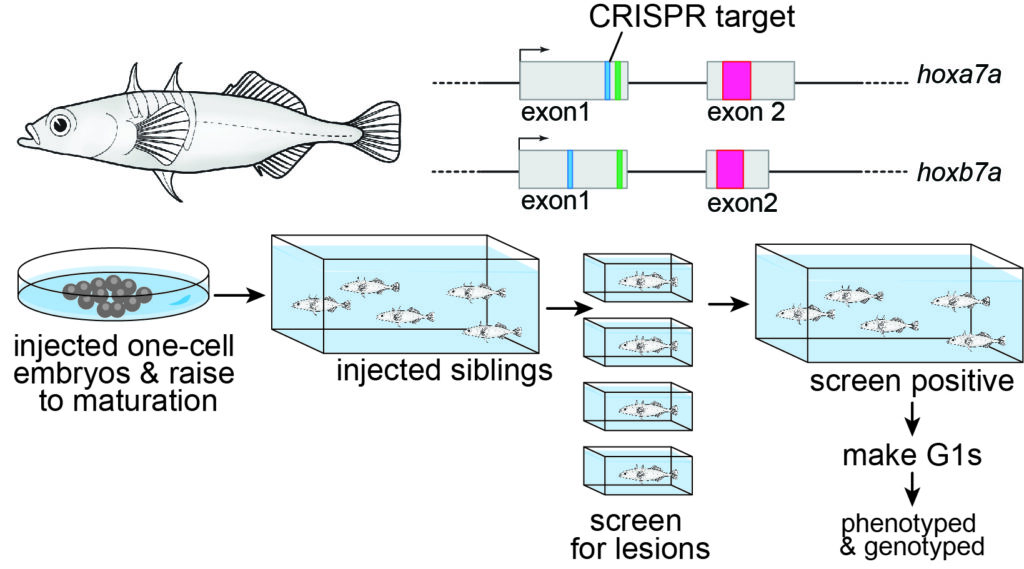
I learned to optimize molecular protocols such as CRISPR/Cas 9 and in situ hybridization, and bioinformatics pipelines for non-model systems, as well as develop skills important for becoming an independent investigator involving scientific writing, experimental design, critical thinking, and public speaking. Based on my findings of unique and convergent losses Hox genomics elements, I designed and executed a CRISPR knockout experiment where I successfully knocked out Hox cluster genes in the stickleback fish. For my doctoral thesis work, I also described a convergent loss of a Hox microRNA and unique loss of a Hox enhancer element and its potential impact on Hox expression in the developing pipefish hindbrain.
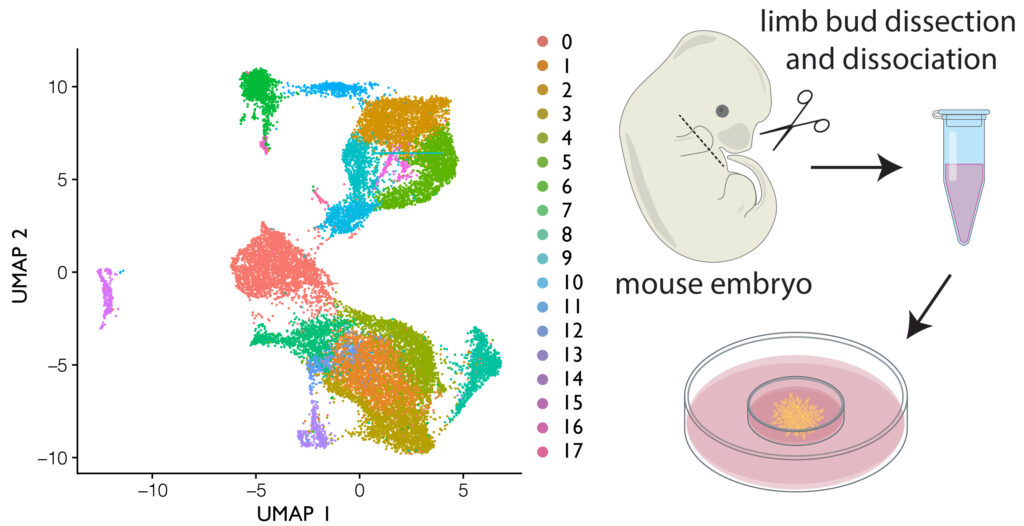
For my current postdoctoral position, I am studying digit development using the distal autopod mesenchyme’s ability to autonomously re-assemble and develop digits, interdigital tissues, and joint structures using a modified micromass assay.
During my time at the University of Oregon, I was fortunate enough to be a teaching assistant for nine different biology courses. I am particularly passionate about teaching evolutionary biology courses, which I have been a teaching assistant for at the University of Kansas and at the University of Oregon, including upper division courses and lab-based courses, which allows me to integrate my personal research interests.