Research
Early research
My early career contributions focused on systematics, taxonomy, and phylogenetics in herpetology. My undergraduate research work at the California Academy of Science involved carefully measuring, sorting, and describing the morphology of preserved museum specimens of Amolops frogs from Myanmar. For my Master’s thesis work at the University of Kansas Biodiversity Institute and Natural History Museum, I participated in a field expedition in the Philippines and I led the publication of the new frog species description. For my thesis, I also described the adaptive osteological traits in two adaptive radiations of island forest frogs. During my time at the University of Kansas, I also worked with a renowned evolutionary biologist, Dr. Ed Wiley, to complete a study that describes the development of the zebrafish caudal skeleton and places it within a phylogenetic context. For that study, I completed fluorescence in situ hybridizations and confocal imaging of a zebrafish developmental series.


Doctoral research
For my doctoral thesis at the University of Oregon, I researched the genomic and development changes that have contributed to the evolution of highly derived morphological characters within pipefish, seahorses, and seadragons. I helped assemble the first genome for this fascinating family of fish and I was the main researcher in charge of annotation the de novo pipefish genome assembly. I described the Hox clusters for the Gulf pipefish (Syngnathus scovelli), which was the first annotated set of Hox genes for syngnathid fish. Based on my findings of unique and convergent losses in Hox genomics elements, I designed and executed a CRISPR knockout experiment where I successfully knocked out Hox cluster genes in the threespine stickleback fish. For my doctoral thesis work, I also described a convergent loss of a Hox microRNA and a unique loss of a Hox enhancer element and its potential impact on Hox expression in the developing pipefish hindbrain.
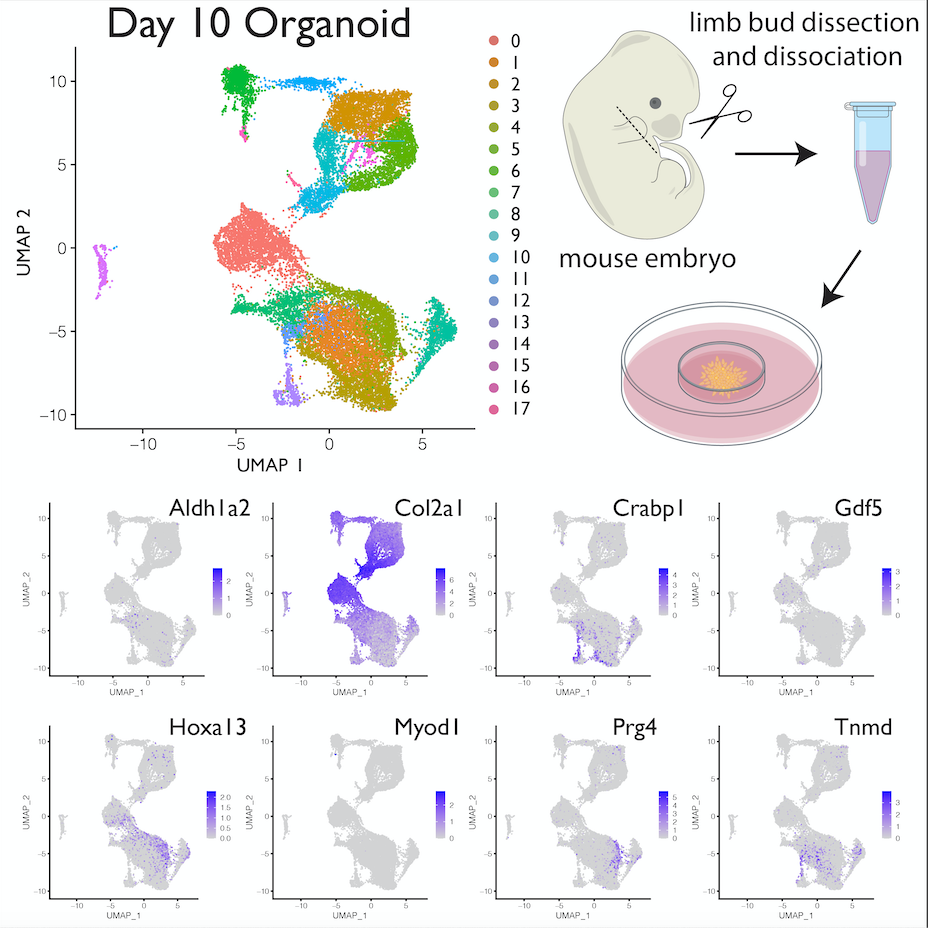
postdoctoral research
For my first postdoctoral fellowship at OHSU, I worked on research projects that focus on single-cell genomics of circulating tumor cells and skin immune cells from patients undergoing immune checkpoint blockade cancer treatments. I helped developed protocols that allow for the isolation of circulating tumor cells from patient blood samples for single-cell RNAseq analysis and I optimized the protocol that processes human skin samples for single-cell analysis. For my second postdoctoral fellowship, I wanted to return to skeletal developmental biology and Hox gene research. In my current research, we have identified a unique property of dissociated distal autopod mesenchyme to autonomously re-assemble and develop into structures resembling digits, interdigital tissues, and joints. This digit organoid system allows us to study digit development outside of the embryo in an in vitro system. For this research, I am doing a single-cell transcriptomic analysis of this digit organoid system to identify cell clusters with significant expression of multiple genes mediating hand/foot development. I am also analyzing the gene expression patterns of signature developmental genes in the digit organoid using HCR RNA-FISH and IHC imaging.